[ad_1]
Database screening
The chemical database was screened based on a 90% similarity index to triclosan and Lipinski’s Rule of five21 revolution to receive a library of 140 compounds. The library is presented in Table S1 with their associated smile structures and PubChem identifiers. This library was chosen as the primary input for extended studies in this report.
Structure‑based pharmacophore modelling
Structure-based e-pharmacophore modelling of the 3D target protein is useful in providing accurate knowledge of protein–ligand interactions39. Default descriptors were used to generate the best e-pharmacophore model. The key contributing residues involved in ligand binding were imported in generating e-pharmacophore of triclosan-FabI complex. The subsequent e-pharmacophore model for A. baumannii FabI protein is characterized with multiple features including H-bond donor, H-bond acceptor, and π–π stacking of aromatic rings as shown in Fig. 1.
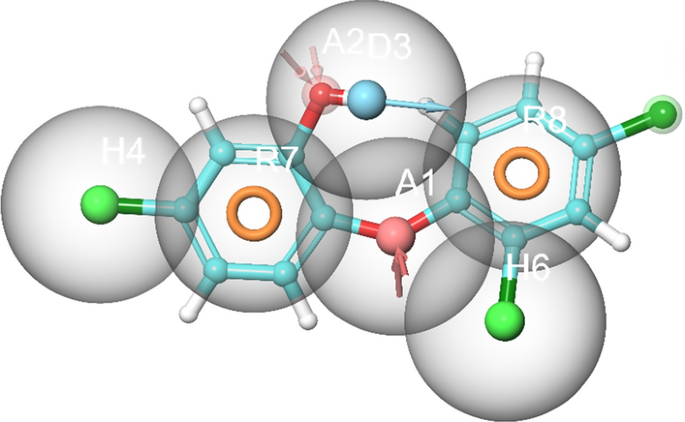
Structure-based 3D pharmacophore modelling based on the analysis of triclosan interactions with the A. baumannii FabI protein and its chemical structure surrounded by excluded volumes. Red arrow: Hydrogen bond acceptor, blue arrow: Hydrogen bond donor, orange sphere: aromatic ring.
Virtual screening of the selected compounds
The attained characteristics of the e-pharmacophore triclosan model in complex with A. baumannii FabI protein were selected as input to filter the prepared library of 140 compounds and are recorded in Table S1. A total of 136 compounds (Table S2) passed the filtering parameters based on the designed e-pharmacophore hypothesis.
Docking‑based virtual screening analysis
The screened 136 compounds were used as input for DBVS as detailed in the methodology. The Glide SP protocol successfully identified 130 compounds (Table S3) following by Glide XP protocol which further narrowed the numbers of compounds to 71 (Table S4) with increased binding scores. These 70 compounds were then optimized to produce one pose per ligand based on the most negative binding score (− 8.03 kcal/mol) against the A. baumannii FabI protein. The top three lead candidate compounds i.e., 21272541, 89795992 and 89792657 underwent ADME/T analysis and these compounds were taken for extended MD simulation studies in process to develop novel inhibitors for A. baumannii infections.
Molecular docking analysis
The top three hit leads (21272541, 89795992, 89792657) with triclosan in complex with the A. baumannii FabI protein were selected for molecular docking studies to check their initial stability in the form of interactions and binding scores. The four 2D structures of model compound triclosan with their 90% similar derivative are presented in Fig. 2.
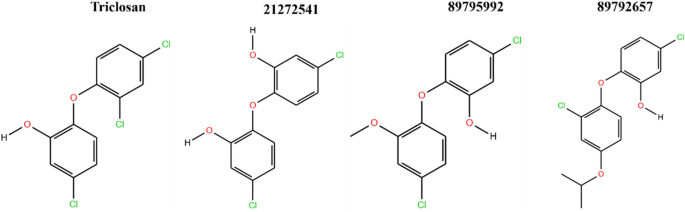
2D structures of model inhibitor triclosan with top three lead compounds.
The active site amino acid residues involved in the binding of selected compounds with A. baumannii FabI protein are shown in Table 1 with their associated binding scores. The binding score for compounds 21272541, 89795992, and 89792657 are − 9.84, − 9.65 and − 9.56 kcal/mol, respectively and were indicative of strong binding with the FabI protein. Furthermore, these scores were higher than the model compound which revealed the binding score of − 8.34 kcal/mol. The binding mode of 21272541 with other two compounds in 3D images and 2D plots are illustrated in Figs. 3 and 4.
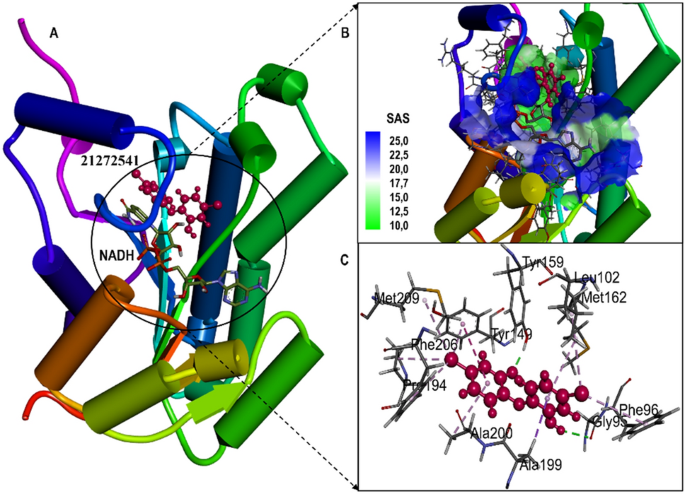
(A) Structural representation of A. baumannii FabI protein in complex with newly developed inhibitor 21272541; (B) Zoomed in view of the binding site residues coloured by solvent accessible surface area of FabI accommodating 21272541 and NADH, (C) Close up view of active residues interacting with the inhibitor showing hydrogen bonds and other Pi-Pi interactions.
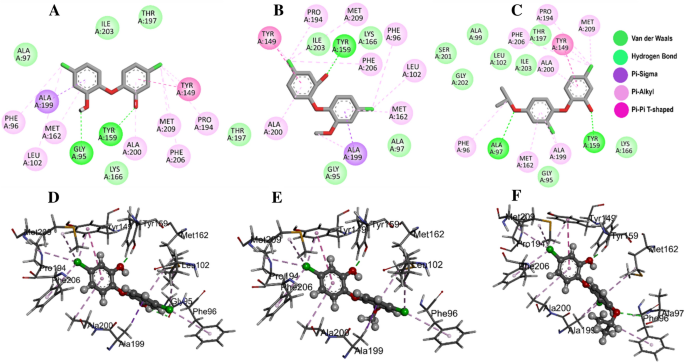
Molecular docking of the FabI protein with the proposed inhibitors: 21272541, 89795992 and 89792657. (A–C) Molecular interactions of the A. baumannii FabI protein with studied compounds with their 2D maps after docking and (D–F) their associated 3D interactions.
Multiple interactions were identified in all the studied compounds (Fig. 4A–C). Compound 21272541 might be the lead inhibitor as it produced two hydrogen bonds with Gly95 and Tyr159. The strong interaction network of the Pi-Sigma was also observed with compound 21272541 and Tyr149 residues which was a key contributing residue in the Klebsiella pneumoniae 21272541-FabI complex in our previous study29. This compound also contributed to the binding by producing Pi-Alkyl interaction which helps in the stability of the complex. The second lead compound 89795992 only formed one hydrogen bond with residue Tyr159 with various Pi-Sigma and Pi-Alkyl networks. While compound 89792657 formed two hydrogen bonds with Ala97 and Tyr159 (Fig. 4D–F). Furthermore, Ala199 was involved in the formation of Pi-Sigma interaction with the benzene ring in all three compounds. The referent inhibitor triclosan only formed one hydrogen bond with Tyr159 and one Pi-Pi stacking with Tyr149 (Figure S1). Tyr159 was thus identified as the key contributor to the binding of A. baumannii FabI protein as it was involved in the hydrogen bond formation of all the three compounds. Nevertheless, compound 21272541 showed the most favourable binding score and was majorly involved in multiple interaction network within the A. baumannii FabI protein and could thus be considered as potential drug candidate after performing extended in-silico studies.
ADME/T analysis
The evaluation of “drug-likeness” properties of the selected compounds with referent inhibitor triclosan were analysed to check if these compounds passed the standard filter criteria. The physicochemical and toxicity characteristics including cell permeation, the influence of metabolism, bioavailability, and toxicity, of the three compounds together with triclosan is detailed in Table 2. We have checked the mentioned characteristics for triclosan with all three drug candidate compounds. Briefly, the projected central nervous system activity (CNS) activities were 1, 0, 1, 1 for triclosan, 21272541, 89795992, and 89792657, respectively and were all in the expected range as mentioned in ADME/T methodology.
Furthermore, the estimated digital values of the human binding serum albumin (QPlogKhsa) of all compounds are in the acceptable range except for triclosan which was − 0.91. The solvent accessible surface area (SASA) of all candidate inhibitors and triclosan was in the acceptable range of 300–1000; however, triclosan had the lowest SASA (356.54) compared to compound 89792657 which exhibited the highest SASA (539.31). Compound 21272541 was identified to have the most suitable octanol/water partition coefficient (QPlogPo/w; (6.16) compared with the other inhibitors which had lower values, albeit still in the expected range of between 2 and 6.5. The aqueous solubility (QPlogS) estimates and the brain/blood partition coefficient (QPlogBB) for all inhibitors with the referent inhibitor were in the acceptable range of − 6.5 to 0.5 and − 3.0 to1.2, respectively. Lastly, all three inhibitors showed 100% human oral absorption (%) except for triclosan which only showed 59.94% human oral absorption and was below the acceptable range (< 25% and > 80%). Nevertheless, candidate compounds pointed towards favorable drug-likeness properties compared to the model inhibitor and could be promising inhibitors of the A. baumannii FabI protein.
Post‑dynamics MD trajectories analysis
The 3D structure of the protein endures major changes after binding of compatible inhibitor which have great effect on proteins biological activity40. In disease effected pathways, the protein-inhibitor binding influences the mode of inhibition of the target protein, thus, analysing the dynamics, fluctuations, and compactness of the protein structure after binding the studied inhibitors was crucial to draw conclusions. After performing 200 ns of MD simulations with selected compounds and triclosan in complex with A. baumannii FabI complex, we analysed the trajectories to observe structural changes. The RMSD of Cα atoms was estimated for 200 ns for all four systems to observe the structural variations in the form of stability and competency.
The obtained RMSD values are plotted in the 2D lot and are sketched in Fig. 5A. All the systems were highly stable till the end of simulation and reached convergence after 50 ns except for the 89792657-FabI system which was unstable and only reached convergence after 150 ns. The 21272541-FabI complex was most stable revealing the RMSD value of 2.23 Å, followed by triclosan, 89792657 and 89795992 complexes with RMSD values of 2.51 Å, 2.89 Å, and 2.91 Å, respectively. Individually, all the systems were measured between 1 and 3.5 Å on the Y axis indicative of stable protein–ligand complexes. This analysis is an evidence that the systems were stable and further investigations performed on these trajectories would be reliable.
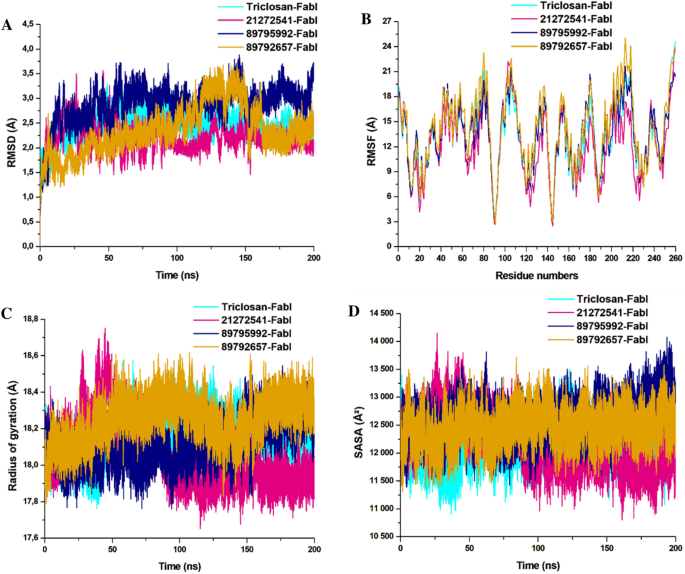
Structural dynamics of A. baumannii FabI protein–ligand complexes. (A) Cα backbone RMSD in Å of all the selected compounds bound to FabI protein; (B) RMSF values after compound binding; (C) RoG value after compound binding; (D) SASA values after compound binding after 200 ns of MD simulations.
Furthermore, the RMSF of Cα atoms were calculated to observe the fluctuations in the amino acids in all four systems. Rigidity and flexibility were measured through this analysis. Complex 21272541-FabI was notably the least fluctuating in comparison with the other two complexes. The 89792657-FabI complex displayed high fluctuation values with 20.01 Å and was higher than the model system as represented in Fig. 5B. This analysis indicates that binding of 21272541 ligand induced least fluctuations in comparison with all other systems and this ligand could be feasible in A. baumannii FabI inhibition.
The RoG analysis was used to validate the folding and unfolding behaviour of the ligand–protein complexes and is crucial to determine the effect of micro molecule entities on the 3D target protein after their binding. The 21272541-FabI complex revealed the least RoG value of 18.02 Å and was stable after 50 ns of simulation time. The 2D plot of RoG can be seen from Fig. 5C. The 89792657-FabI complex revealed the highest RoG value of 18.34 Å indicative of less compactness and folding activities. All four systems; however, were between 17.81 and 18.5 Å values. Triclosan and 89795992-FabI complexes followed similar patterns of compactness. Better compactness and improved binding of selected compounds, specifically to 21272541 was offered with this analysis against A. baumannii FabI protein.
The last structural analysis included SASA which was performed to determine the hydrophobic and hydrophilic effects of the amino acids exposed to the solvent molecules during the MD simulations (Fig. 5D). The SASA values for all the systems were from 11,110 to 13,701 Å2. The 21272541-FabI complex showed the lowest SASA value and was also less exposed to solvents in comparison to other complexes. Furthermore, the triclosan-FabI complex was also similar to the 21272541-FabI complex; however, was slightly more exposed to the solvent compared with 89792657-FabI complex. Nevertheless, the alterations in the SASA values among the three selected compounds with model inhibitor are correlated with the folding and unfolding of the A. baumannii protein. Binding of the 21272541 compound is an indication of improved exposure to solvents and enhanced inhibition activity towards the FabI protein. The retention of all three ligands within the binding site of FabI protein was confirmed by generating snapshots initially at 10 ns and afterwards at every 50 ns till 200 ns of MD simulation as shown in Figure S2.
Intramolecular hydrogen bond analysis
The intramolecular hydrogen bond analysis is essential in estimating the conformation and stability of the protein. This evaluation extensively infers the mechanism of ligand–protein binding with better understanding in terms of hydrogen bond formation. The overall number of intramolecular hydrogen bonds in the 21272541-FabI complex were noted from 86 to 149 whereas in 89795992-FabI complex, it showed between 87 and 145. Minor differences were observed in these two complexes (Fig. 6), including a lower intramolecular hydrogen bonds of compound 89795992 compared to the triclosan-FabI complex. This intramolecular hydrogen bond analysis supports the above structural findings observed on A. baumannii FabI protein in complex with top three hits.
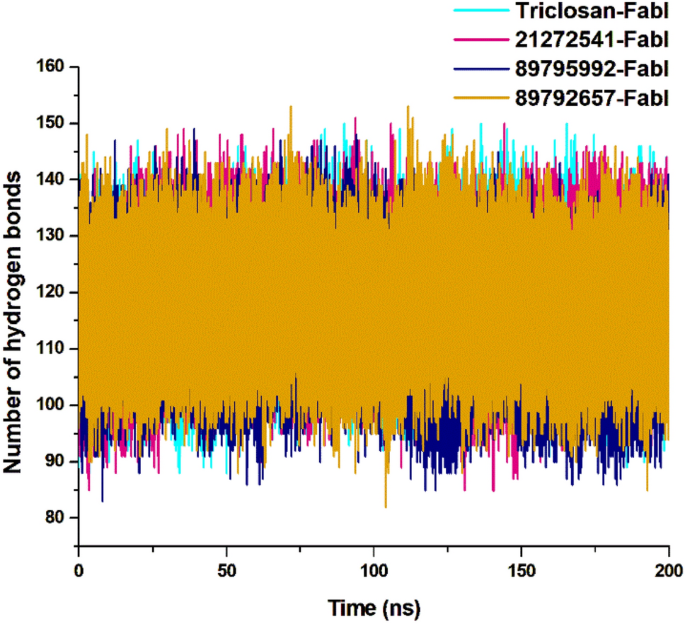
Intramolecular hydrogen bond analysis for studied complexes, calculated after 200 ns MD simulation.
Distance correlation matrix analysis
The dynamics of the three selected compounds with triclosan in complex with the FabI protein were calculated by performing dynamic cross correlation matrix (DCCM) for the negative and positive residual correlation movements as shown in Fig. 7. The A. baumannii FabI protein can be seen dispersed into various positive and negative communities throughout the simulation. Triclosan-FabI, 89795992-FabI, and 89792657-FabI complexes followed a similar pattern of scattering; however, significant changes were noted in 21272541-FabI complex with mostly positive residual movement between the amino acids. Furthermore, the highly anti-correlated regions of the compounds range from 200 to 353 amino acids as they are not directly involved in the binding of selected ligands which could indicate lower inhibitory activity. Thus, this analysis suggests that the amino acids within the FabI protein are in positive correlation to 21272541 and this compound will most likely reduce the inhibition of the FabI protein.
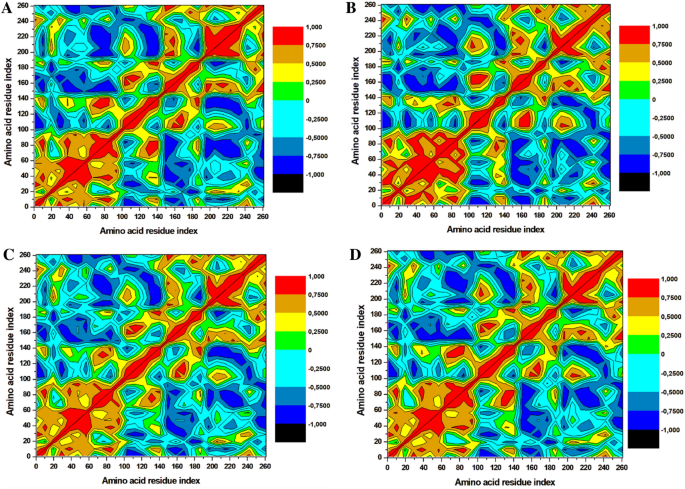
Dynamics cross-correlation matrix analysis. (A) Triclosan-FabI and (B) 21272541-FabI (C) 89795992-FabI and (D) 89792657-FabI complex calculated after 200 ns of MD trajectories.
Principal component analysis
The principal component analysis (PCA) was performed for the assessment of conformational modifications in amino acids upon ligand binding based on eigenvectors on the X and Y axis29. The continuous scattering of ligands in the PC1–PC2 points can be noted in the 2D plot in Fig. 8. Triclosan scattered in negative positions on PC1 whereas all three selected compounds are scattered on positive positions on PC1–PC2 eigenvectors. The 3D porcupine plot for all four systems was produced to interpret the ligands movements in various directions as displayed in Fig. 8. The compound 21272541 exhibited the most positive movements on eigenvector 1 on X axis; however, on the Y axis it showed a negative movement with a negative trace covariance matrix. It also displayed the trace covariance matrix of 20.18 Å with positive values on X axis. Compounds 89795992 and 89792657 denoted the positive correlation with least fluctuation on eigenvector 1 and 2 with positive values after binding to A. baumannii FabI protein. This analysis supports the previously mentioned DCCM analysis for positive and negative residual movements of complexes throughout the MD simulation.
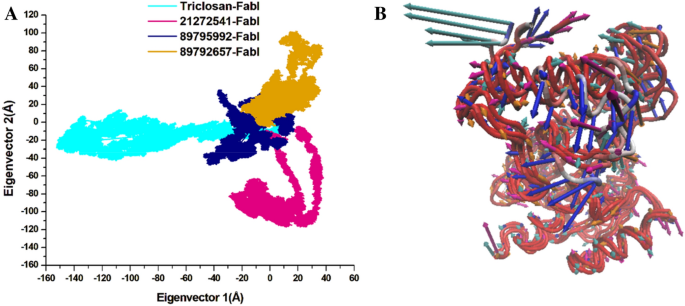
(A) PCA plot constructed by eigenvector 1 versus eigenvector 2 for the studied compounds with triclosan; (B) Porcupine plot of PC1 collective motions for the obtained predominant eigenvectors over the 200 ns MD trajectories for triclosan (cyan colour), 21272541 (pink colour), 89795992 and (blue colour), 89792657 (orange colour) and in complex with FabI protein.
Binding affinity calculation with MM/GBSA
The overall binding affinities of various components of energies including electrostatic, van der Waal, polar, non-polar, solvation, and gas-phase energy were calculated with MM/GBSA technique. It is well-validated method to estimate the free energies involved in the binding of chemical molecules to biological macromolecules based on the MD simulation studies. The outcome from this analysis is recorded in Table 3. The 21272541-FabI complex revealed the most favourable free binding energy of − 61.63 kcal/mol as expected and was the most accurate compound with all the structural, hydrogen bond and PCA analysis. The model triclosan-FabI complex also showed better binding of − 59.02 kcal/mol in comparison with the 89795992-FabI and the 89792657-FabI complexes which exhibited the binding energies of − 52.09 kcal/mol and − 38.29 kcal/mol, respectively. The van der Waals energies was higher in the 89795992-FabI complex whereas it was lower in the 89792657-FabI with the digital values of − 46.68 kcal/mol and − 34.16 kcal/mol, respectively. Furthermore, the electrostatic energy was the highest in the 21272541-FabI complex whereas it was the lowest in 89795992-FabI complex with major difference of − 17.30 kcal/mol detected between the two compounds. The overall gas phase energies were favourable for all the studied complexes. The electrostatic and van der Waals energies are the most important contributors in ligand protein binding and they all showed promising results. The total binding energy is the difference of polar and non-polar energies and was accurately calculated as seen in the tabular results. The findings from all components of energies are indicative of significant binding of compound 21272541 to FabI protein and further validates this ligand to have the capacity to inhibit the activity of A. baumannii FabI protein and could potentially overcome drug resistant mechanisms.
Per-residue energy decomposition analysis with MM/GBSA
The per-residue energy decomposition approach is commonly used to calculate the energy contribution of each amino acid and their association with the target protein in assisting rational of drug design. A cluster of 15 common amino acids were selected based on their involvement in the binding of 21272541, 89795992 and 89792657 compounds and model drug triclosan with A. baumannii FabI protein.
Tyr149 and Tyr159 are key residues in the 21272541 binding with the most promising energies of − 1.81 kcal/mol and − 1.73 kcal/mol, respectively. Furthermore, these residues were involved in the formation of hydrogen bonds and Pi-Alkyl interactions with each studied ligands (Fig. 9). Residue Ile203 also showed better binding energy of 21272541 with value of − 1.84 kcal/mol compared to referent inhibitor with value of − 1.54 kcal/mol; however, this residue did not produce any interaction network. The least contributing amino acids were from model drug triclosan exhibiting lower residual energies as compared to our screened compounds. Thus, from this analysis we observed the contribution of specific residues involved in strong binding.
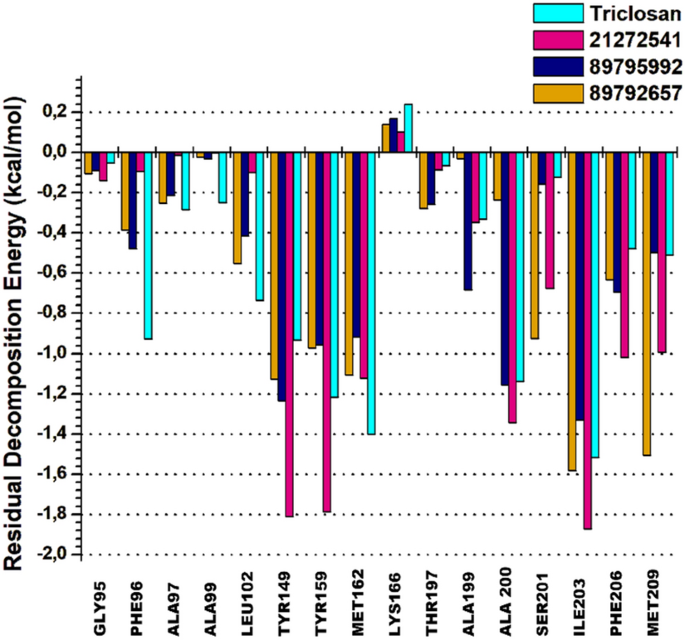
2D plot of per-residual energy decomposition of A. baumannii FabI protein in complex with the studied compounds and model drug triclosan.
[ad_2]
Source link
